Degrader Building Blocks
Tocris provides Building Blocks to support Degrader research and development. Degraders (e.g. PROTAC® molecules, SNIPERs etc) are heterobifunctional small molecules that are modular in design, comprising binding moieties for an E3 ubiquitin ligase and a target protein joined by a linker. Our Degrader components have functional handles for easy conjugation to ligands/linkers of interest. The range includes the most effective and commonly used E3 ubiquitin ligase ligands, functionalized at positions known not to interfere with binding affinity. E3 ligase ligands conjugated to common linker groups are also supplied. Below you will find a toolbox of Degrader Building Blocks to support targeted protein degradation research and development. You can also select your own panel of E3 ligase ligands + linkers for your Degrader development program using our new PROTAC Panel Builder tool.
PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
VHL Targeting Ligands Plus Linkers
| Cat. No. | Product Name | Structure | E3 Ligase Exit Vector | Linker
Type |
Number
of Units |
Conjugation
Functionality |
|---|---|---|---|---|---|---|
| 7106 | VH 032 amide-alkylC2-acid |

|
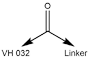 |
Alkyl | 2 | Carboxylic Acid |
| 6852 | VH 032 amide-alkylC2-amine |

|
 |
Alkyl | 2 | Amine |
| 7216 | VH 032 amide-alkylC3-acid |

|
 |
Alkyl | 3 | Acid |
| 7073 | VH 032 amide-alkylC3-amine |

|
 |
Alkyl | 3 | Amine |
| 6680 | VH 032 amide-alkylC4-acid |

|
 |
Alkyl | 4 | Carboxylic Acid |
| 6465 | VH 032 amide-alkylC4-amine |

|
 |
Alkyl | 4 | Amine |
| 7217 | VH 032 amide-alkylC5-acid |

|
 |
Alkyl | 5 | Acid |
| 6985 | VH 032 amide-alkylC5-amine |

|
 |
Alkyl | 5 | Amine |
| 7867 | VH 032 amide-alkylC5-azide |

|
 |
Alkyl | 5 | Azide |
| 7107 | VH 032 amide-alkylC6-acid |

|
 |
Alkyl | 6 | Carboxylic Acid |
| 6853 | VH 032 amide-alkylC6-amine |

|
 |
Alkyl | 6 | Amine |
| 7925 | VH 032 amide-alkylC7-acid |

|
 |
Alkyl | 7 | Carboxylic Acid |
| 7074 | VH 032 amide-alkylC7-amine |

|
 |
Alkyl | 7 | Amine |
| 7108 | VH 032 amide-alkylC8-acid |

|
 |
Alkyl | 8 | Carboxylic Acid |
| 6890 | VH 032 amide-alkylC8-amine |

|
 |
Alkyl | 8 | Amine |
| 7404 | VH 032 amide-alkylC9-acid |

|
 |
Alkyl | 9 | Carboxylic Acid |
| 7566 | VH 032 amide-alkylC9-amine |

|
 |
Alkyl | 9 | Amine |
| 6880 | VH 032 amide-alkylC10-amine |

|
 |
Alkyl | 10 | Amine |
| 7104 | VH 032 amide-PEG1-acid |

|
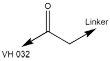 |
PEG | 1 | Acid |
| 6879 | VH 032 amide-PEG1-amine |

|
 |
PEG | 1 | Amine |
| 6907 | VH 032 amide-PEG2-amine |

|
 |
PEG | 2 | Amine |
| 6684 | VH 032 amide-PEG2-alkyne |

|
 |
PEG | 2 | Alkyne |
| 6679 | VH 032 amide-PEG3-acid |

|
 |
PEG | 3 | Carboxylic Acid |
| 6463 | VH 032 amide-PEG3-amine |

|
 |
PEG | 3 | Amine |
| 7215 | VH 032 amide-PEG4-acid |

|
 |
PEG | 4 | Carboxylic Acid |
| 6464 | VH 032 amide-PEG4-amine |

|
 |
PEG | 4 | Amine |
| 7105 | VH 032 amide-PEG5-acid |

|
 |
PEG | 5 | Acid |
| 7568 | VH 032 amide-PEG6-amine |

|
 |
PEG | 6 | Amine |
| 6909 | VH 032 phenol-alkylC4-amine |

|
 |
Alkyl | 4 | Amine |
| 6910 | VH 032 phenol-alkylC6-amine |

|
 |
Alkyl | 6 | Amine |
| 7293 | VH 101 phenol-alkylC4-amine |

|
 |
Alkyl | 4 | Amine |
Cereblon Targeting Ligands Plus Linkers
| Cat. No. | Product Name | Structure | E3 Ligase Exit Vector | Linker
Type |
Number
of Units |
Conjugation
Functionality |
|---|---|---|---|---|---|---|
| 7516 | Lenalidomide 4'-alkylC3-azide |

|
- | alkyl | 3 | Azide |
| 7517 | Lenalidomide 4'-alkylC5-azide |

|
- | alkyl | 5 | Azide |
| 7075 | Lenalidomide 4'-PEG1-amine |

|
- | PEG | 1 | Amine |
| 7513 | Lenalidomide 4'-PEG1-azide |

|
- | PEG | 1 | Azide |
| 7093 | Lenalidomide 4'-PEG2-amine |

|
- | PEG | 2 | Amine |
| 7514 | Lenalidomide 4'-PEG2-azide |

|
- | PEG | 2 | Azide |
| 7076 | Lenalidomide 4'-PEG3-amine |

|
- | PEG | 3 | Amine |
| 7515 | Lenalidomide 4'-PEG3-azide |

|
- | PEG | 3 | Azide |
| 7472 | Lenalidomide 5'-piperazine |

|
Piperazine | - | - | Amine |
| 7473 | Lenalidomide 5'-piperazine-4-methylpiperidine |

|
Piperazine | Methyl-piperidine | 1 | Amine |
| 7998 | PAG 4'-piperazine |

|
Piperazine | - | - | Amine |
| 7999 | PAG 4'-piperazine-4-methylpiperidine |

|
Piperazine | Methyl-piperidine | 1 | Amine |
| 6851 | Pomalidomide 4'-alkylC2-amine |

|
- | Alkyl | 2 | Amine |
| 7439 | Pomalidomide 4'-alkylC2-azide |

|
- | Alkyl | 2 | Azide |
| 7096 | Pomalidomide 4'-alkylC3-acid |

|
- | Alkyl | 3 | Carboxylic Acid |
| 7205 | Pomalidomide 4'-alkylC3-amine |

|
- | Alkyl | 3 | Amine |
| 7440 | Pomalidomide 4'-alkylC3-azide |

|
- | Alkyl | 3 | Azide |
| 7209 | Pomalidomide 4'-alkylC4-acid |

|
- | Alkyl | 4 | Carboxylic Acid |
| 7441 | Pomalidomide 4'-alkylC4-azide |

|
- | Alkyl | 4 | Azide |
| 6682 | Pomalidomide 4'-alkylC5-acid |

|
- | Alkyl | 5 | Carboxylic Acid |
| 6716 | Pomalidomide 4'-alkylC5-amine |

|
- | Alkyl | 5 | Amine |
| 7442 | Pomalidomide 4'-alkylC5-azide |

|
- | Alkyl | 5 | Azide |
| 7210 | Pomalidomide 4'-alkylC6-acid |

|
- | Alkyl | 6 | Carboxylic Acid |
| 7923 | Pomalidomide 4'-alkylC6-amine |

|
- | Alkyl | 6 | Amine |
| 7443 | Pomalidomide 4'-alkylC6-azide |

|
- | Alkyl | 6 | Azide |
| 7097 | Pomalidomide 4'-alkylC7-acid |

|
- | Alkyl | 7 | Carboxylic Acid |
| 7208 | Pomalidomide 4'-alkylC7-amine |

|
- | Alkyl | 7 | Amine |
| 7211 | Pomalidomide 4'-alkylC8-acid |

|
- | Alkyl | 8 | Carboxylic Acid |
| 6944 | Pomalidomide 4'-alkylC8-amine |

|
- | Alkyl | 8 | Amine |
| 7732 | Pomalidomide 4'-alkylC10-amine |

|
- | Alkyl | 10 | Amine |
| 7094 | Pomalidomide 4'-PEG1-acid |

|
- | PEG | 1 | Acid |
| 7203 | Pomalidomide 4'-PEG1-amine |

|
- | PEG | 1 | Ethylamine |
| 7434 | Pomalidomide 4'-PEG1-azide |

|
- | PEG | 1 | Azide |
| 7212 | Pomalidomide 4'-PEG2-acid |

|
- | PEG | 2 | Acid |
| 6637 | Pomalidomide 4'-PEG2-amine |

|
- | PEG | 2 | Amine |
| 7307 | Pomalidomide 4'-PEG2-azide |

|
- | PEG | 2 | Azide |
| 6681 | Pomalidomide 4'-PEG3-acid |

|
- | PEG | 3 | Carboxylic Acid |
| 7204 | Pomalidomide 4'-PEG3-amine |

|
- | PEG | 3 | Ethylamine |
| 7435 | Pomalidomide 4'-PEG3-azide |

|
- | PEG | 3 | Azide |
| 7213 | Pomalidomide 4'-PEG4-acid |

|
- | PEG | 4 | Acid |
| 6963 | Pomalidomide 4'-PEG4-amine |

|
- | PEG | 4 | Ethylamine |
| 7436 | Pomalidomide 4'-PEG4-azide |

|
- | PEG | 4 | Azide |
| 7095 | Pomalidomide 4'-PEG5-acid |

|
- | PEG | 5 | Acid |
| 7437 | Pomalidomide 4'-PEG5-azide |

|
- | PEG | 5 | Azide |
| 7438 | Pomalidomide 4'-PEG6-azide |

|
- | PEG | 6 | Azide |
| 7468 | Pomalidomide 5'-piperazine |

|
Piperazine | - | - | Amine |
| 7469 | Pomalidomide 5'-piperazine-4-methylpiperidine |

|
Piperazine | Methyl-piperidine | 1 | Amine |
| 7470 | Pomalidomide 5'-fluoro-6-piperazine |

|
Piperazine | - | - | Amine |
| 7471 | Pomalidomide 5'-fluoro-6-piperazine-4-methylpiperidine |

|
Piperazine | Methyl-piperidine | 1 | Amine |
| 7329 | Thalidomide 4'-ether-alkylC2-acid |

|
 |
Alkyl | 2 | Acid |
| 6850 | Thalidomide 4'-ether-alkylC2-amine |

|
 |
Alkyl | 2 | Amine |
| 7330 | Thalidomide 4'-ether-alkylC3-acid |

|
 |
Alkyl | 3 | Acid |
| 7521 | Thalidomide 4'-ether-alkylC3-azide |

|
 |
Alkyl | 3 | Azide |
| 7331 | Thalidomide 4'-ether-alkylC4-acid |

|
 |
Alkyl | 4 | Acid |
| 6917 | Thalidomide 4'-ether-alkylC4-amine |

|
 |
Alkyl | 4 | Amine |
| 7332 | Thalidomide 4'-ether-alkylC5-acid |

|
 |
Alkyl | 5 | Acid |
| 7522 | Thalidomide 4'-ether-alkylC5-azide |

|
 |
Alkyl | 5 | Azide |
| 6627 | Thalidomide 4'-ether-alkylC6-amine |

|
 |
Alkyl | 6 | Amine |
| 7333 | Thalidomide 4'-ether-alkylC6-acid |

|
 |
Alkyl | 6 | Acid |
| 7829 | Thalidomide 4'-ether-alkylC6-azide |

|
 |
Alkyl | 6 | Azide |
| 7334 | Thalidomide 4'-ether-alkylC7-acid |

|
 |
Alkyl | 7 | Acid |
| 7335 | Thalidomide 4'-ether-alkylC8-acid |

|
 |
Alkyl | 8 | Acid |
| 7403 | Thalidomide 4'-ether-alkylC8-amine |

|
 |
Alkyl | 8 | Amine |
| 7518 | Thalidomide 4'-ether-PEG1-azide |

|
 |
PEG | 1 | Azide |
| 6686 | Thalidomide 4'-ether-PEG2-alkyne |

|
 |
PEG | 2 | Alkyne |
| 7519 | Thalidomide 4'-ether-PEG2-azide |

|
 |
PEG | 2 | Azide |
| 6950 | Thalidomide 4'-ether-PEG3-amine |

|
 |
PEG | 3 | Ethylamine |
| 7520 | Thalidomide 4'-ether-PEG3-azide |

|
 |
PEG | 3 | Azide |
| 6951 | Thalidomide 4'-ether-PEG5-amine |

|
 |
PEG | 5 | Ethylamine |
| 6949 | Thalidomide 4'-oxyacetamide-alkylC2-amine |

|
 |
Alkyl | 2 | Amine |
| 6469 | Thalidomide 4'-oxyacetamide-alkylC4-amine |

|
 |
Alkyl | 4 | Amine |
| 6968 | Thalidomide 4'-oxyacetamide-alkylC6-amine |

|
 |
Alkyl | 6 | Amine |
| 6946 | Thalidomide 4'-oxyacetamide-alkylC8-amine |

|
 |
Alkyl | 2 | Amine |
| 6300 | Thalidomide 4'-oxyacetamide-alkylC4-azide |

|
 |
Alkyl | 4 | Azide |
| 6966 | Thalidomide 4'-oxyacetamide-PEG1-amine |

|
 |
PEG | 1 | Ethylamine |
| 6967 | Thalidomide 4'-oxyacetamide-PEG2-amine |

|
 |
PEG | 2 | Ethylamine |
| 6467 | Thalidomide 4'-oxyacetamide-PEG3-amine |

|
 |
PEG | 3 | Amine |
| 6468 | Thalidomide 4'-oxyacetamide-PEG4-amine |

|
 |
PEG | 4 | Amine |
| 6913 | Thalidomide 4'-oxyacetamide-alkylC1-PEG3-alkylC3-amine |

|
 |
Alkyl-PEG-Alkyl | 1-3-3 | Amine |
| 7988 | Thalidomide 5'-amine-alkylc6-amine |

|
 |
Alkyl | 6 | Ethylamine |
| 7557 | Thalidomide 5'-amine-alkylc8-amine |

|
 |
Alkyl | 8 | Ethylamine |
| 7733 | Thalidomide 5'-amine-alkylC10-amine |

|
 |
Alkyl | 10 | Ethylamine |
| 7735 | Thalidomide 5'-amine-PEG1-amine |

|
 |
alkyl | 10 | Ethylamine |
| 7736 | Thalidomide 5'-amine-PEG2-amine |

|
 |
PEG | 2 | Ethylamine |
| 7737 | Thalidomide 5'-amine-PEG3-amine |

|
 |
PEG | 3 | Ethylamine |
IAP Targeting Ligands Plus Linkers
| Cat. No. | Product Name | Structure | E3 Ligase Exit Vector | Linker
Type |
Number
of Units |
Conjugation
Functionality |
|---|---|---|---|---|---|---|
| 6474 | A 410099.1 amide-alkylC4-amine |

|
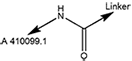 |
Amide-Alkyl | 4 | Amine |
| 6473 | A 410099.1 amide-PEG2-amine |

|
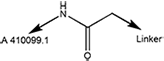 |
Amide-PEG | 2 | Amine |
| 6472 | A 410099.1 amide-PEG3-amine |

|
 |
Amide-PEG | 3 | Amine |
| 7220 | A 410099.1 amide-PEG4-amine |

|
 |
Amide-PEG | 4 | Amine |
| 7221 | A 410099.1 amide-PEG5-amine |

|
 |
Amide-PEG | 5 | Amine |
Functionalized E3 Ligase Ligands
| Cat. No. | Product Name | Structure | E3 Ligase Ligand | Target E3 | Reactive Handle | Reaction Chemistry |
|---|---|---|---|---|---|---|
| 8066 | CST 530, Phenol |

|
CST 530 | IAP | Alcohol | Mitsunobu reaction |
| 6471 | A 410099.1, amine |

|
A 410099.1 | IAP | Primary Amine | Amide coupling |
| 7178 | LCL 161, phenol |

|
LCL 161 | IAP | Alcohol | Mitsunobu reaction |
| 7677 | Lenalidomide 5'-amine |

|
Lenalidomide | Cereblon | Primary Amine | Amide Coupling |
| 7858 | PD 4'-oxyacetic acid |

|
Phenyl Dihydrouracil | Cereblon | Carboxylic Acid | Amide Coupling |
| 7859 | PD 4'-piperazine |

|
Phenyl Dihydrouracil | Cereblon | Cyclic Amine | Amide Coupling |
| 7670 | Phenyl-glutarimide 4'-oxyacetic acid |

|
Phenyl-glutarimide | Cereblon | Carboxylic Acid | Amide Coupling |
| 6466 | TC E3 5031 |

|
Thalidomide | Cereblon | Carboxylic acid | Amide coupling |
| 6628 | TC E3 5032 |

|
Pomalidomide | Cereblon | Fluorine | Nucleophilic aromatic substitution |
| 7456 | Thalidomide, 5'-fluoro |

|
Thalidomide | Cereblon | Fluoro | Nucleophilic aromatic substitution |
| 6685 | Thalidomide, propargyl |

|
Thalidomide | Cereblon | Alkyne | Click chemistry |
| 6462 | VH 032, amine |

|
VH 032 | VHL | Primary amine | Amide coupling |
| 6969 | cis VH 032, amine dihydrochloride |

|
VH 032 | VHL | Primary amine | Amide coupling |
| 7634 | Me VH 032, amine |

|
VH 032 | VHL | Primary amine | Amide coupling |
| 6911 | VH 032, phenol |

|
VH 032 | VHL | Alcohol | Mitsunobu reaction |
| 6683 | VH 032, propargyl |

|
VH 032 | VHL | Alkyne | Click chemistry |
| 7861 | VH 032, thiol |

|
VH 032 | VHL | Thiol | Nucleophilic substitution |
| 6952 | VH 101, phenol |

|
VH 101 | VHL | Alcohol | Mitsunobu reaction |
| 7884 | VH 101, thiol |

|
VH 101 | VHL | Thiol | Nucleophilic substitution |
Functionalized Warhead Ligands
| Cat. No. | Product Name | Structure | Ligand | Target Protein | Reactive Handle | Reaction Chemistry |
|---|---|---|---|---|---|---|
| 7689 | (+)-JQ1 bump, acid functionalized |

|
JQ1 analog | BET Bromodomains | Carboxylic acid | Amide coupling |
| 6588 | (+)-JQ1 carboxylic acid |

|
JQ1 | BET Bromodomains | Carboxylic acid | Amide coupling |
| 7576 | (+)-JQ1 maleimide |

|
JQ1 | BET Bromodomains | Maleimide | Sulfhydryl coupling |
| 6589 | (+)-JQ1 PA |

|
JQ1 | BET Bromodomains | Alkyne | Click chemistry |
Require a different derivative? Get in touch.
PROTAC is a registered trademark of Arvinas Operations, Inc., and is used under license.
Literature for Degrader Building Blocks
Tocris offers the following scientific literature for Degrader Building Blocks to showcase our products. We invite you to request* your copy today!
*Please note that Tocris will only send literature to established scientific business / institute addresses.
TPD and Induced Proximity Research Product Guide
This brochure highlights the tools and services available from Bio-Techne to support your Targeted Protein Degradation and Induced Proximity research, including:
- Active Degraders
- TAG Degradation Platform
- Degrader Building Blocks
- Assays for Protein Degradation
- Induced Proximity Tools
Developing Degraders Poster
This poster describes the generation of a database of Degraders (PROTACs®) from the literature. The Degraders were profiled according to the constituent ligands, linker type, linker length and physicochemical properties and this information was used to establish a set of guidelines for the design and synthesis of cell-permeable Degrader molecules. Presented at the 20th SCI/RSC Medicinal Chemistry Symposium 2019, Cambridge, UK.PROTAC® is a registered trademark of Arvinas Operations, Inc., and is used under license.
Enabling Research By Provision of Chemical Tools Poster
The Tocris chemistry team has considerable chemistry knowledge and skill which it has recently been applying to the generation of new chemical probes from known tools and compounds to help answer biological questions. This poster presents some of the work carried out by Tocris scientists and focuses on two areas: the generation of libraries of chemical building blocks to support Degrader (PROTAC) research and the development of a fluorescent probe to study integrin biology. Presented at Chemical Tools for Complex Biological Systems II, 2019, Janelia Research Campus, USA.Targeted Protein Degradation Poster
Degraders (e.g. PROTACs) are bifunctional small molecules, that harness the Ubiquitin Proteasome System (UPS) to selectively degrade target proteins within cells. They consist of three covalently linked components: an E3 ubiquitin ligase ligand, a linker and a ligand for the target protein of interest. Authored in-house, this poster outlines the generation of a toolbox of building blocks for the development of Degraders. The characteristics and selection of each of these components are discussed. Presented at EFMC 2018, Ljubljana, Slovenia



